Hey i am working on my Bachelor thesis at the moment and use UNET in combination with a GAN for image segmentation. I spend the last 5 months on that, so on my tests, the new approach of januar 2020, called Multires-UNET is quite a good choice for more texture orientated segmentation. I use the current github implementation. Its quite nice, maybe you notice that you can easyly tweak the number of parameters with "alpha" in the implementation, to scale the multiple Resnets in the Unet structure.
I tried also other segmentation networks like Mask_RCNN with different backbones or tried to construct various types of CAE on my own, but always had to come back to a UNET like structure. Same goes for ResU-NET and R2U, the multires one worked better for my purposes cause i didnt need any kind of LSTM modules.
Some examples which may clarify the difference in performance on a specific task:
Original Image:
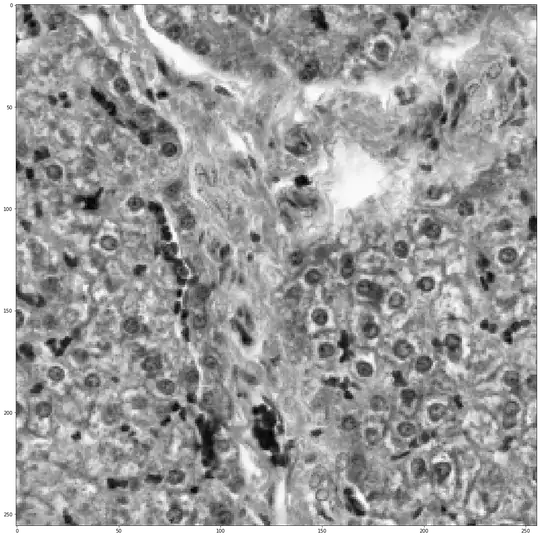
Ground-Truth:

Classical UNET++ (Unet with skip-connections)(2.5 million parameters) more parameters (wide) didnt change the result.
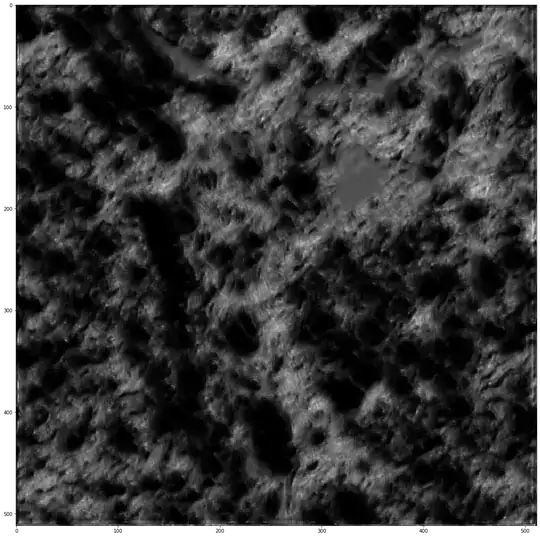
Multires-UNET (alpha=1.67, think was about 7 million parameters)

Can you show some of ur microscopy images, how complex is ur task and what do you want to segment ?



