I am working on image segmentation of MRI thigh images with deep learning (Unet). I noticed that I get a higher average dice accuracy over my predicted masks if I have less samples in the test data set. I am calculating it in tensorflow as
def dice_coefficient(y_true, y_pred, smooth=0.00001):
y_true_f = K.flatten(y_true)
y_pred_f = K.flatten(y_pred)
intersection = K.sum(y_true_f * y_pred_f)
return (2. * intersection + smooth) / (K.sum(y_true_f) + K.sum(y_pred_f) + smooth)
the difference is 0.003 if I have 4x more samples.
I am calculating the dice coefficient over each MRI 2D slice
Why could this be?
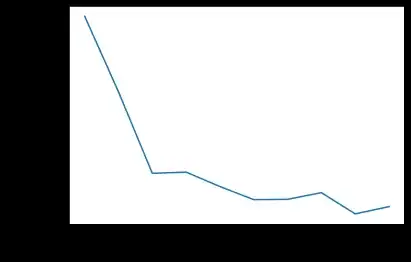
This figure shows how the accuracy decreases with the fraction of samples. I start with 0.1 of the data until the whole data set. The splitting of the data was random