You can use mmv (install it with sudo apt install mmv) as follows:
mmv -n 'paired_dat_*2_R2*' 'paired_dat_#1\1_R2#2'
-n is used so that you can preview the changes without applying them to your files. If you are satisfied with the output, run the command without -n.
How this works
mmv comprises of a filename matching part ('paired_dat_*2_R2*' here) and a replacement part ('paired_dat_#1\1_R2#2' here).
In the first part we use wildcards to match parts of the filename which allows for using these matches to rename the files.
In the above command the * wildcard is used, which means "match any character". The first * matches the characters between paired_dat_ and 2_R2, while the second * matches the characters from 2_R2 to the end of the filename.
Each match can be then used in the replacement part using # followed by ascending numbers that correspond to the match. So #1 corresponds to the match of the first * and #2 to the second match of *. Since the first * matches up to 2 without including it, using #1 in the replacement part essentially removes it. We then add 1_R2 (1 should be escaped, thus we use \1) and then we add the rest of the filename as #2.
You can find more details by running in a terminal man mmv.
There is also a GUI option using Nautilus, which is similar to what Artur Meinild suggests in his answer:
Open Nautilus.
Navigate to the directory with your files.
Select your files.
right-click and select Rename or press F2.
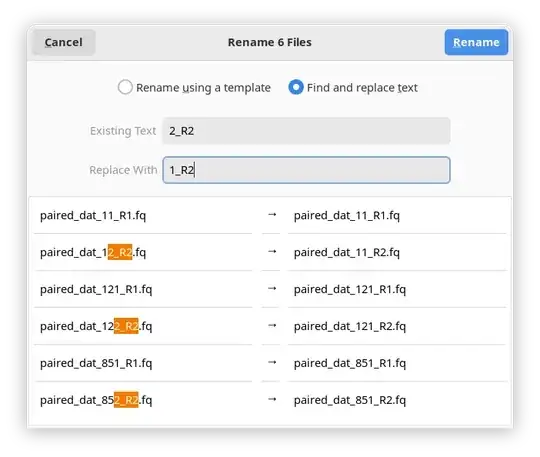
In the window that pops up select Find and replace text.
In the Existing Text field enter: 2_R2
In the Replace With field enter: 1_R2
Click Replace and your files will be renamed!
As always, test this first on a copied portion of the files to make sure that it works as intended.

12to11? Change2to1? Change2to its previous number? [Edit] your question and be more specific. Also add a few more items to your example input and output formats so we can understand better. – BeastOfCaerbannog Sep 01 '22 at 08:532to1so the ID number is the same while leaving theR1andR2alone since those correspond to forward and reverse ends of the genome. – veridian21 Sep 01 '22 at 10:03